Our Projects
1. Mapping the microbial communities across the Indian Coast line using 16s metagenomics
GD Khedkar, BA Chopade, Mahesh Solunkhe, Amol Kalyankar, Dinesh Nalage, Bharathi Prakash, Sanil Pillai Amrin Fatima
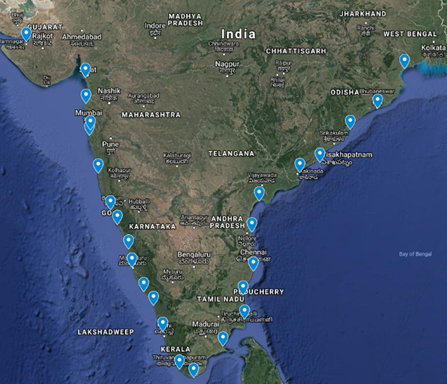
The key questions to be explored;
- Overall microbial community known (%) unknown (%)
- East coast vs west coast diversity disparities
- Influence of rivers on microbial communities
- Effects of ports on microbial communities
- Effect of human populations (Mumbai, Surat, Goa, Kochin, Kanyakumati, Vijawada, Puri, Kakinada etc)
2. Mapping the marine fish species of India
(GD Khedkar, BA Chopade, Anita Tiknaik, Amol Kalyankar, Ishwarya Deepty, Mamatha Dalala, Ganesh Korhale, Nazneen Deshmukh, Bharathi Prakash, Biju Kumar, Persis Manne, A Chandrashekhar Reddy)
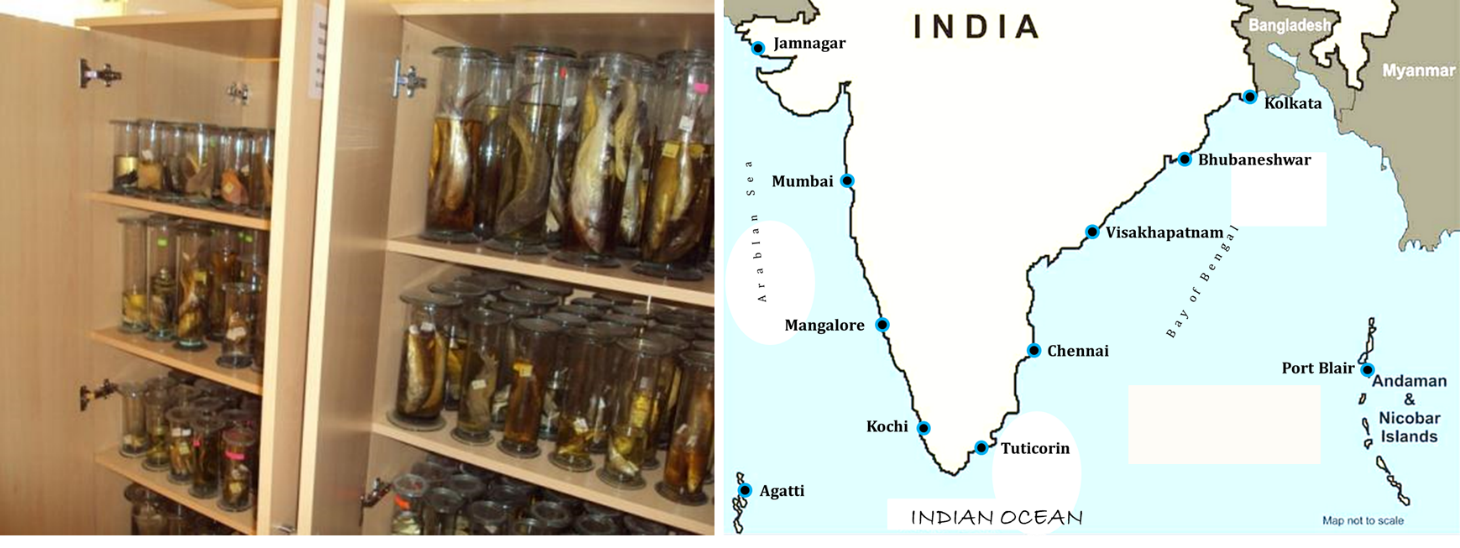
The key questions to be explored;
- Marine fish species diversity mapping using DNA Barcoding
- Resolving taxonomic ambiguities
- Diversity disparities in terms of genetic distance
- Application in commercial practices
3. Mapping the Indian rivers for fish diversity
(GD Khedkar, Sandeep Rathod, Vikas Kalyankar, Anita Tiknaik, Amol Kalyankar, Mahesh Shingare, Sanil Pillai, Ganesh Korhale, Mahesh Shingare, Rahul Suryawanshi, Deepali Sangale, Tejashwini Sontakke, A Chandrashekhar Reddy, Lior David)
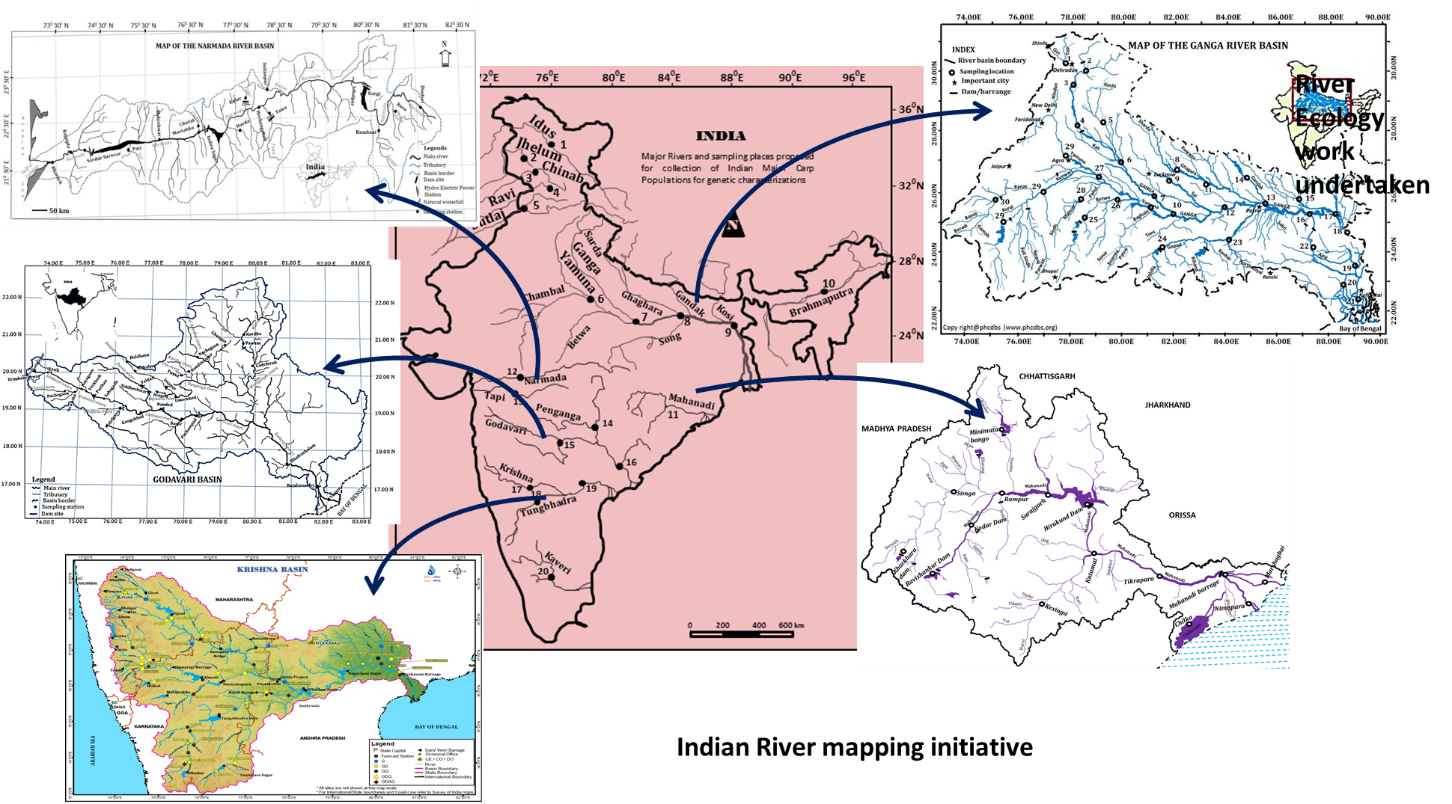
The key questions to be explored;
- Marine fish species diversity of major river basins in India
- Resolving taxonomic ambiguities
- Comparing trans-basin diversity status
- Application in conservation programs
4. Wildlife Forensics
(GD Khedkar, Dinesh Nalage, Anil Sarkate, Anita Tiknaik, Amol Kalyankar, Sanil Pillai, Ganesh Korhale, Deepali Sangale, Tejashwini Sontakke, Raituja Hange, Shil Abayankar, Vikram Khilare)
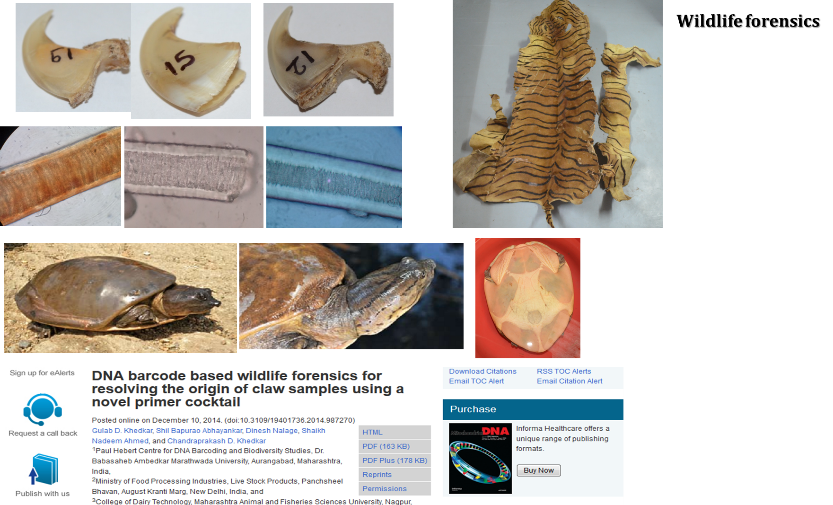
The key questions to be explored;
- Develop DNA forensics methods for various wildlife species in India
- Assist forest department in such cases
- Assist in conservation programes
5. Whole genome sequencing project (20 whole genomes)
(GD Khedkar, BA Chopade, Dinesh Nalage, Anil Sarkate, Anita Tiknaik, Amol Kalyankar, Sanil Pillai, Rajshri Deolalikar, Deepali Sangale, Tejashwini Sontakke, Bharathi Prakash, CD Khedkar, Atsushi kurabayashi, P. Raju, Mamatha Dadla)

The key questions to be explored;
- Generate genome map of important species in India for higher productivity, species Improvement and conservation
- Disease research
- Generate a lead in such type of research
- Assist farmers, fisher society
6. Whole mitochondrial genome project (500 whole genomes)
(GD Khedkar, BA Chopade, Dinesh Nalage, Anil Sarkate, Anita Tiknaik, Amol Kalyankar, Sanil Pillai, Chandrakant Jadhav, Vipin Hiremath, Rajshri Deolalikar, Ganesh Korhale, Mahesh Shingare, Rahul Suryawanshi, Deepali Sangale, Tejashwini Sontakke, Bharathi Prakash, CD Khedkar, Atsushi kurabayashi, Mamatha Dadla, P. Raju)
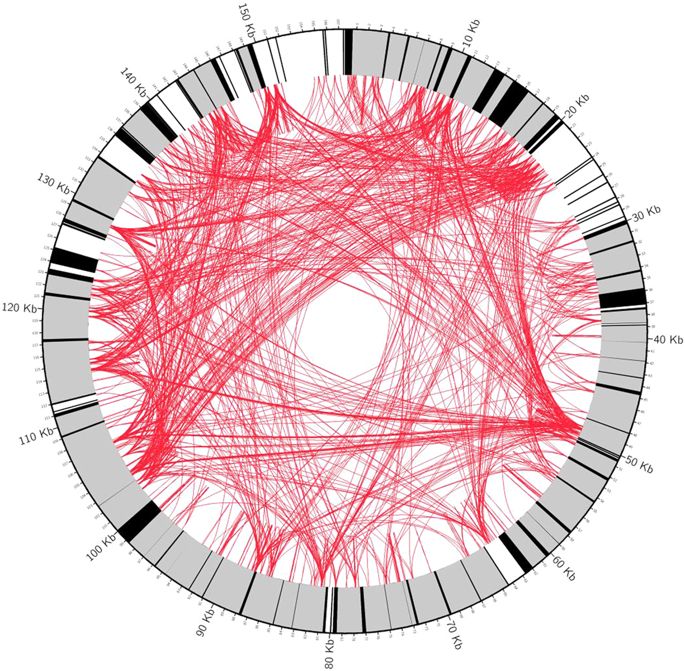
The key questions to be explored;
- Generate mitochondrial genome map of important species in India to unwind the history of their evolution
- Built phylogenetic history of every species
- Comparative account of their mt. genomes
7. Comparative account of vertebrate gut microbiota (36 organisms including tiger, elephant)
(GD Khedkar, BA Chopade, Dinesh Nalage, Anil Sarkate, Anita Tiknaik, Amol Kalyankar, Sanil Pillai, Rajshri Deolalikar,Ganesh Korhale, Deepali Sangale, Tejashwini Sontakke, Bharathi Prakash, CD Khedkar, Atsushi Kurabayashi, P. Raju, Mamatha Dadla)
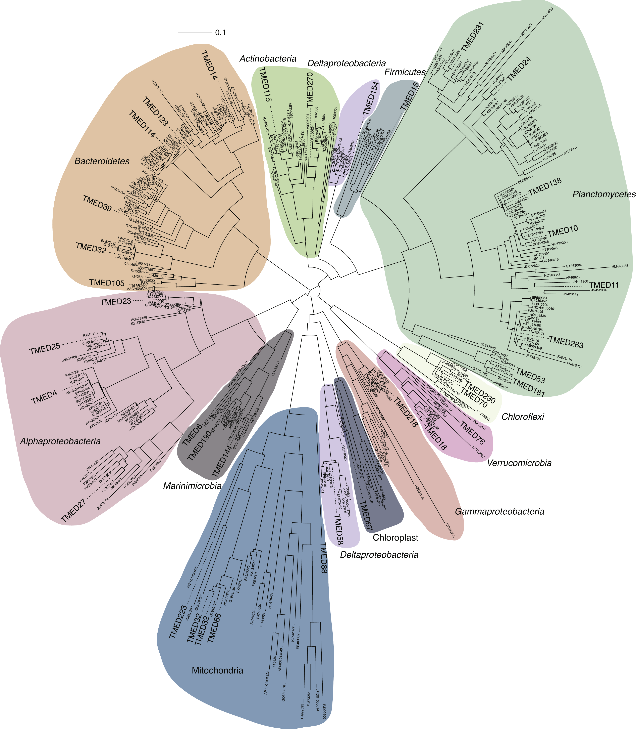
The key questions to be explored;
- 1. Generate genome map gut microbiome of various vertebrate species
- 2. Correlation between diet and microbial community
- 3. Possibilities for correlational studies
- 4. Assist farmers, fisher society
8. Transcriptomic analysis of thermos tolerance silkworm breeds so as to plan breeding for better hybrids (500 transcriptomes)
(GD Khedkar, Raju P., Mamatha Dadla, Anita Tiknaik, Swetha, Vaibhav Late,)
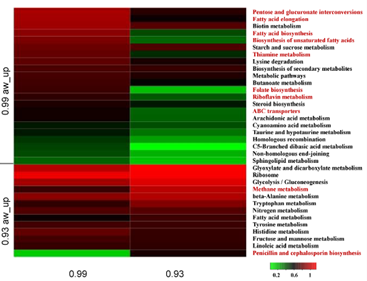
The key questions to be explored;
- Challenge with thermal treatments and Profiling corresponding gene expression
- Target specific genes for thermal tolerance
- Analysis for heterozygosity of such genes and built breeding program